Research
Summary
Translation quality controls eliminate aberrant proteins to maintain protein homeostasis and normal cell function. Improving the accuracy of translation and preventing the production of abnormal proteins is a practical approach for suppressing a series of neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease. We analyze the molecular mechanism of quality control mechanisms that suppress abnormal proteins and clarify drug discovery’s molecular basis. We propose that the increase in translation accuracy and the enhancement of translation quality control mechanisms are possible strategies to prevent abnormal protein production and prolong healthy life expectancy. We have been analyzing the molecular mechanism of quality control mechanisms that suppress the production of abnormal proteins to clarify the molecular basis of drug discovery (Figure 1). We reported that co-translational degradation of the nascent peptide by ribosome stalling by poly(A) sequence and tandem rare codons (Inada and Aiba, EMBO J., 2005; Ito-Harashima et al., Genes Dev. 2007; Dimitrova et al., JBC, 2009; Kuroha et al., EMBO Rep. 2009). It leads to the discovery of Ribosome-associated Quality Control (RQC) that monitors aberrant translations and decomposes and removes abnormal proteins during synthesis. Recent studies in the last ten years have elucidated its molecular mechanism and physiological function. We also reported a molecular mechanism that recognizes and dissociates stagnant ribosomes during translation elongation, the early stage of RQC (Kuroha et al., EMBO Rep. 2009, 2010; Tsuboi et al., Mol. Cell, 2012; Izawa et al., Cell Rep., 2012; Tsuboi et al., JBC, 2015; Ikeuchi et al., Sci. Rep., 2016; Figure 2). In the last several years, we have reported that E3 ubiquitin ligase Hel2 and its mammalian homolog ZNF598 are required for RQC, and the novel RQT complex is involved in the dissociation of the ubiquitinated ribosomes to subunits (Matsuo et. al., Nat. Communi. 2017). Our group and Hegde’s lab have reported that E3 ubiquitin ligase recognizes collided ribosomes (Disome/Trisome), and the specific structure of collided ribosomes. We reported that the ubiquitination of uS10 by Hel2 was reconstituted (Ikeuchi et al., EMBO J., 2019; Matsuo et al., NSMB, 2020). Next, we identified an RQT complex that specifically dissociates ubiquitinated ribosomes into subunits and reconstituted the reaction in vitro (Matsuo et al., NSMB, 2020; Hashimoto et al., Sci. Rep. 2020). We also elucidated the structures of ANKZF1 and 60S ribosomes that cleave peptidyl-tRNA on 60S (Su, Izawa et al., Nature, 2019). The collision of ribosomes also induces NGD (No-Go Decay) quality controls in conjugation with RQC and triggers endonucleolytic cleavage of mRNA in the collided ribosome (Ikeuchi et al., EMBO J., 2019; Figure 2). We also reported two pathways of NGD, mRNA cleavage coupled to the dissociation of collided ribosome response in RQC and mRNA cleavage independent of RQC at the vicinity of the collided ribosomes (Ikeuchi et al., EMBO J., 2019). We also reported that molecular mechanism of quality control NRD for deficient ribosomes (Sugiyama, Li, Kato et.al., Cell Rep., 2019; Figure 3), the function of ribosome dynamic modification in stress response (Matsuki et al., BBRC 2020; Matsuki, Matsuo et al., Sci. Rep. 2020), and discovered new molecular mechanisms that determine mRNA stability (Buschauer, Matsuo et al., Science, 2020; Figures 4-6).
It has become clear that translation quality controls play critical roles as the very early stage of maintaining protein homeostasis. Suppose the increase in the translation accuracy and the translation quality control mechanism could be a possible strategy to prevent abnormal protein production and prolong healthy life expectancy.
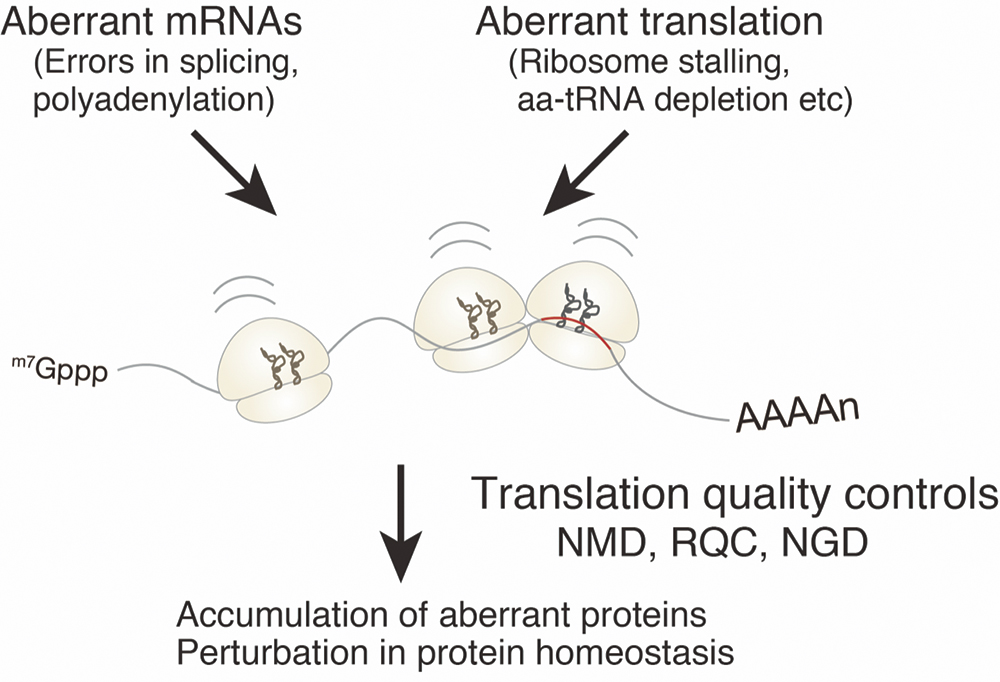
Figure 1. Accumulation of aberrant proteins in cells causes various cell dysfunctions. Translation quality controls eliminate and prevent aberrant proteins from maintaining protein homeostasis normal cell function.
Quality control for translation abnormalities Molecular mechanism of RQC and NGD
Ribosome-associated Quality Control (RQC) monitors aberrant translations and decomposes and removes abnormal proteins during synthesis (Bengtson and Joazeiro, Nature 2010; Brandman et al., Cell 2012). RQC plays an extremely important role as the very early stage of maintaining protein homeostasis. We have reported a molecular mechanism that recognizes and dissociates stagnant ribosomes during translation elongation, the early stage of RQC (Kuroha et al., EMBO Rep. 2009, 2010; Tsuboi et al., Mol. Cell, 2012; Izawa et al., Cell Rep., 2012; Tsuboi et al., JBC, 2015; Ikeuchi et al., Sci. Rep., 2016).
In the last several years, we have reported that E3 ubiquitin ligase Hel2 and its mammalian homolog ZNF598 are required for RQC, and the novel RQT complex is involved in the dissociation of the ubiquitinated ribosomes to subunits (Matsuo et. al., Nat. Communi. 2017). We and Hegde lab have reported that E3 ubiquitin ligase recognizes collided ribosomes (Disome/Trisome), and the specific structure of collided ribosomes (Figure 2). We reported that the ubiquitination of uS10 by Hel2 was reconstituted (Ikeuchi et al., EMBO J., 2019; Matsuo et al., NSMB, 2020). Next, we identified an RQT complex that specifically dissociates ubiquitinated ribosomes into subunits and reconstituted the reaction in vitro (Matsuo et al., NSMB, 2020; Hashimoto et al., Sci. Rep. 2020). We also elucidated the structures of ANKZF1 and 60S ribosomes that cleave peptidyl-tRNA on 60S (Su, Izawa et al., Nature, 2019). The collision of ribosomes also induces NGD (No-Go Decay) quality controls in conjugation with RQC and triggers endonucleolytic cleavage of mRNA in the collided ribosome (Ikeuchi et al., EMBO J., 2019; Figure 2). We also reported two pathways of NGD, mRNA cleavage coupled to the dissociation of collided ribosome response in RQC and mRNA cleavage independent of RQC at the vicinity of the collided ribosomes (Ikeuchi et al., EMBO J., 2019).
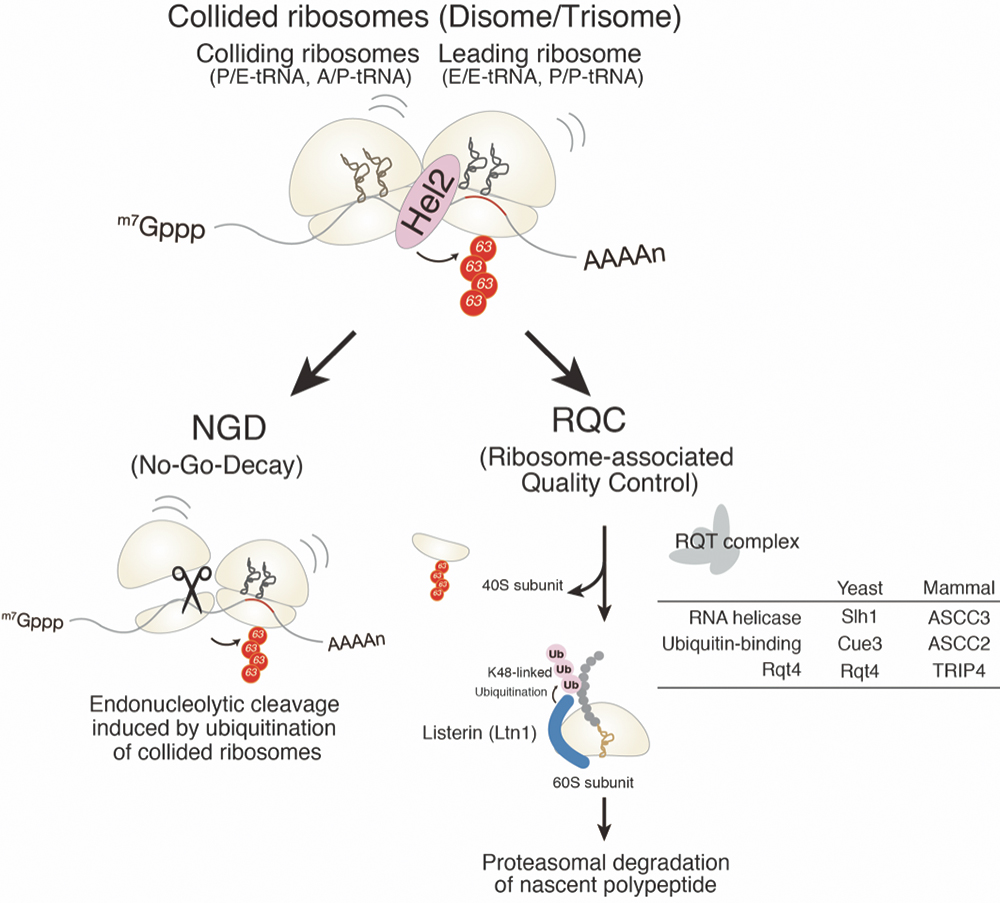
Figure 2. Quality controls recognize aberrant translation and eliminate its aberrant products.
Molecular mechanism of quality control NRD for deficient ribosomes
The ribosome is the central machinery for protein synthesis responsible for accurate codon recognition and highly efficient peptide bond formation. The ribosome interacts with various factors to perform essential functions for gene expression. Since abnormal ribosomes generated during the synthesis cause various expression abnormalities, cells have a quality control mechanism Nonfunctional Ribosomal RNA Decay (NRD) recognizes and eliminates functionally defective ribosomes. We recently analyzed the quality control of ribosomes deficient in function due to base substitution mutations conserved in all species, essential for accurate codon recognition in 18S rRNA, and ubiquitin at the K212 residue of ribosomal protein uS3. We identified E3 ubiquitin ligases that are essential and involved (Sugiyama, Li, Kato et.al., Cell Rep., 2019; Figure 3). It was also revealed that the ubiquitinated stagnant 80S ribosome was dissociated into each subunit by Slh1, and then the abnormal 40S was degraded.
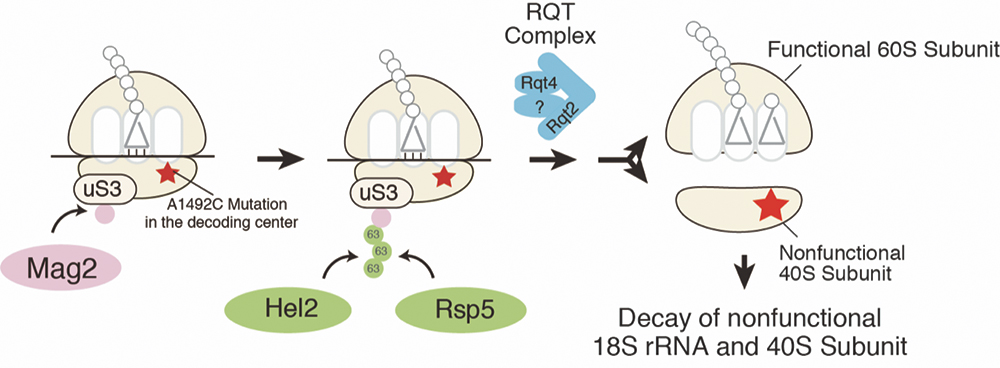
Figure 3. E3 ubiquitin ligases recognize and ubiquitinate stalled ribosomes to eliminate nonfunctional 40S subunit and 18S rRNA.
The function of ribosome dynamic modification in stress response
Synthesis and modification of secretory proteins in the endoplasmic reticulum are essential for cells. Accumulation of abnormal proteins in the endoplasmic reticulum is harmful to cells and therefore responds by inducing the UPR pathway. In Saccharomyces cerevisiae, the membrane protein Ire1 activated by endoplasmic reticulum stress splices the precursor mRNA of the transcription factor Hac1, and Hac1 is synthesized to induce transcription of chaperones. In higher eukaryotes, PARK phosphorylates eIF2α and suppresses cell-wide translation initiation. In the process of elucidating the physiological function of ribosomal ubiquitination, we discovered a novel translational regulation in the endoplasmic reticulum stress response. We discovered a novel translational control mechanism during endoplasmic reticulum stress in S. cerevisiae and clarified that ubiquitination of the ribosomal protein eS7 by E3 ubiquitin ligase Not4 is essential (Matsuki et al., BBRC 2020; Matsuki, Matsuo et al., Sci. Rep. 2020).
Discovery of new molecular mechanisms that determine mRNA stability
The genetic information encoded by mRNA is converted into protein by the ribosome. The ribosome decodes the genetic information using the three base sequences on the mRNA as one reading frame (codon), but multiple types of codons encode most amino acids. Each codon corresponding to the same amino acid is called a synonymous codon, and the intracellular abundance of the corresponding tRNA is biased. The codons that are often used are called optimal codons, and whether synonymous codons are optimal is quantified according to the abundance of the corresponding tRNA, and the higher the amount of tRNA, the higher the optimality. Therefore, it is known that when translating mRNA with many codons with optimal codons, the elongation rate is high, and many proteins are synthesized. Codon optimality plays a crucial role in gene expression because the translation elongation rate is closely linked to the regulation of expression level and the folding and targeting of the peptide chain to be synthesized. In recent years, Coller lab reported that the optimality of codons regulates the inherent stability of individual mRNAs (Presnyak et al., Cell, 2015). It has been established as a general rule that mRNAs with more optimal codons are stable and mRNAs with less optimal codons are unstable. The higher the optimal codon, the faster the translation elongation rate, so the optimal codon determines the mRNA’s half-life. On the other hand, the mechanism by which the rate of translation elongation regulated by codon optimality is monitored and individual mRNAs’ inherent stability remains still unclear.
We have elucidated the molecular basis by which Ccr4-Not senses the translation elongation rate and determines the degradation rate as an mRNA stability-determining mechanism, in collaboration with Beckmann lab and Coller lab (Buschauer, Matsuo et al., Science, 2020). It is widely known that the Ccr4-Not complex involved in transcription/degradation and translational repression of mRNA binds to mRNA via RNA-binding protein, but we first used biochemical techniques to Ccr4. Buschauer in Beckmann lab has found that the Not5 subunit of the Ccr4-Not complex binds directly to the ribosome. Subsequently, a comprehensive analysis by selective ribosome profiling was performed to characterize the mRNA translated by the ribosome to which the Ccr4-Not complex specifically binds. The results showed that codon-level analysis showed that the affinity between the Ccr4-Not complex and the ribosome showed a very strong inverse correlation with codon optimality.
We also showed that the Ccr4-Not complex dysfunction results in the loss of mRNA stability control by codon optimization. In other words, stabilization caused by mRNA with high codon optimization (many optimal codons) and destabilization caused by mRNA with low codon optimization (fewer optimal codons) is eliminated. These clarified that the Ccr4-Not complex leads to mRNA degradation by having a strong affinity for ribosomes that translate mRNA with low codon optimization (Figure 4). Next, we determined the structure of the Ccr4-Not complex bound to the ribosome. Single-particle analysis using a cryo-electron microscope revealed that Not5, one of the constituent proteins of the Ccr4-Not complex, binds to the ribosome’s E site, which does not contain tRNA at the A site (Figure 5). During the translation process, codon-anticodon recognition is performed by the A site of the ribosome. Ribosomes translating non-optimal codons have a low abundance of corresponding tRNAs, so it takes a long time for the tRNAs to bind to the A site. Since tRNA does not bind to site A, tRNA dissociates from site E. It was found that the Ccr4-Not complex binds to the E site where tRNA is dissociated and emptied, leading to efficient mRNA degradation (Figure 6). From the above results, it is clear that the Ccr4-Not complex monitors codon optimality and controls mRNA stability by efficiently binding non-optimal codons to the ribosome’s E site during translation.
This study clarified the actual state of the mRNA degradation control mechanism by codon optimization and the significance of synonymous codons in genetic information that had been unknown for many years. Abnormal translation rate regulation during protein synthesis causes severe defects in protein function, leading to disruption of protein homeostasis. Proteostasis disruption causes a wide range of cellular dysfunctions such as defective protein accumulation, organelle damage, and disruption of signaling pathways. It is thought to cause neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease, and aging. These results are expected to serve as a basis for understanding the pathogenic mechanism and aging mechanism of diseases caused by the synthesis of function-deficient proteins due to translation abnormalities.
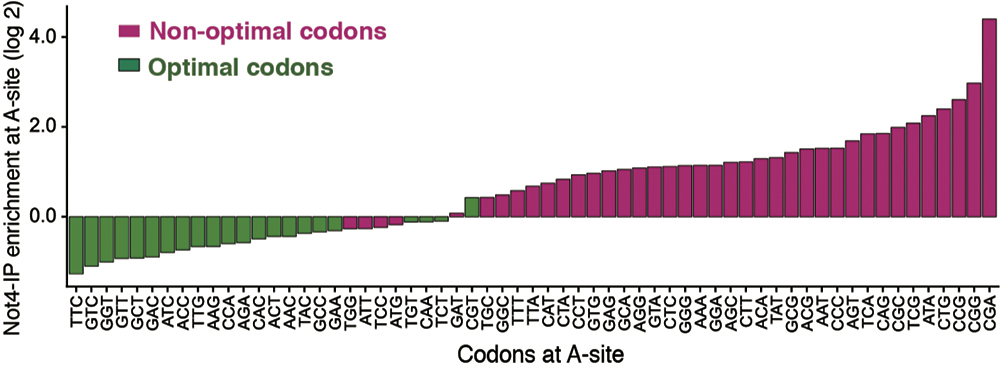
Figure 4. The affinity between Ccr4-Not and the ribosome shows a strong inverse correlation with the optimal codon. The vertical axis shows the affinity of the Ccr4-Not complex for the ribosome, and the horizontal axis shows the codon at the A site of the ribosome. Ribosomes containing the non-optimal codon (red) showed high affinity for the Ccr4-Not complex, and ribosomes containing the optimal codon (green) showed low affinity. Therefore, it was revealed that a less optimal codon exists at the ribosome’s A site to which the Ccr4-Not complex binds.
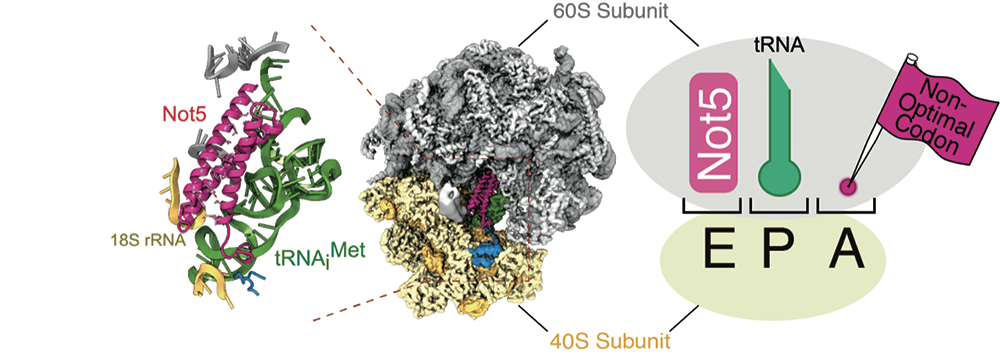
Figure 5. Not5 and ribosome binding mode. Left figure: Single-particle analysis of Ccr4-Not complex and ribosome containing no tRNA at site A using cryo-electron microscopy. Not5, one of the constituent proteins of the Ccr4-Not complex, binds to the E site of the ribosome. Pink indicates the amino-terminal region of Not5, green indicates tRNA, yellow indicates the 40S ribosomal subunit and gray indicates the 60S ribosomal subunit. Right figure: When a non-optimal codon is present at the A site of the ribosome, Not5 binds to the E site, causing mRNA degradation.
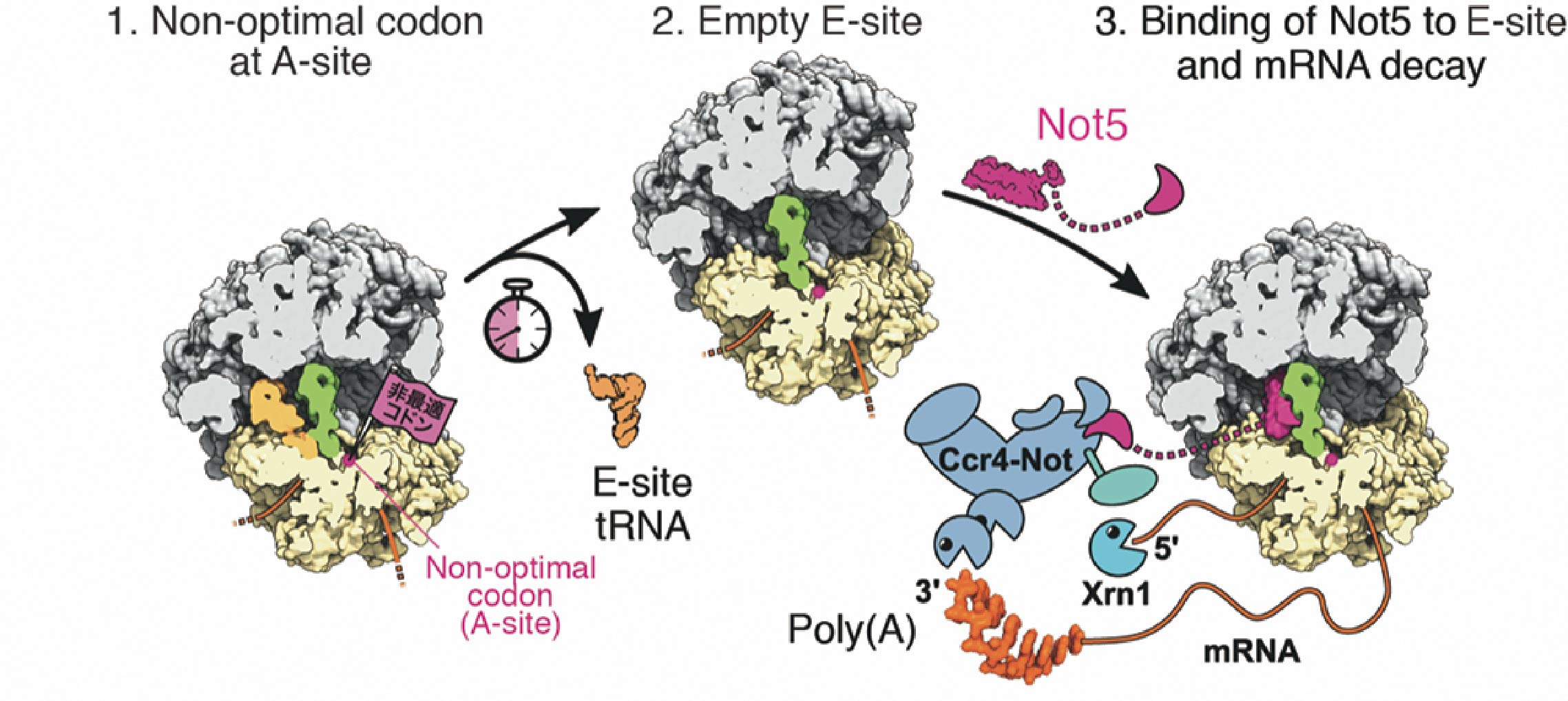
Figure 6. mRNA degradation control mechanism model depending on codon optimality (1) A ribosome in which the non-optimal codon is located at the A site. Due to the low abundance of tRNA, it takes a long time for tRNA to bind to the A site. (2) Since tRNA does not bind to site A, tRNA dissociates from site E. (3) The N-terminal region of the Not5 subunit of the Ccr4-Not complex binds to the E site. The Ccr4-Not complex bound to the ribosome shortens poly(A). Also, ribonuclease Xrn1 degrades mRNA after removal of the cap structure. Pink indicates the amino-terminal region of Not5. Magenta indicates tRNA bound to the E site, green indicates tRNA bound to the P site, yellow indicates the 40S ribosomal subunit, gray indicates the 60S ribosomal subunit, and orange indicates mRNA.